一种重组双功能融合蛋白及应用
文献发布时间:2023-06-19 10:36:57
技术领域
本发明属于融合蛋白技术领域,具体涉及一种重组双功能融合蛋白及应用。
背景技术
间皮素(mesotnelin,MSLN)是一种40kDa的细胞表面糖蛋白,不是癌症特异性抗原,是一种分化抗原,在正常组织中以相对较低的水平表达。与正常组织相比,间皮素在多种类型的实体瘤中高表达,例如胰腺癌、卵巢癌、恶性间皮瘤、肺腺癌以及子宫内膜癌、胆道胃癌和前列腺癌。间皮素在胸膜、心包和腹膜内衬的正常间皮细胞上表达。间皮素在正常组织上的有限分布使其成为肿瘤特异性治疗的有望靶标。间皮素基因编码一个71kDa的前体蛋白,由2138bp长的cDNA编码,包含628个氨基酸,该蛋白被加工成一个40kDa的膜结合蛋白(称为间皮素)和一个31kDa的脱落片段,称为巨核细胞增强因子(megakaryocyte-potentiating factor,MPF),该片段从细胞中释放出来。间皮素蛋白是单克隆抗体K1识别的抗原,通常是与膜结合的糖脂酰肌醇(GPI)连接的蛋白质;而MPF是从人胰腺癌细胞系的培养基中分离得到的。间皮素在约30%的癌症中过表达,肿瘤间皮素的表达通常与肿瘤侵袭性增加和临床不良预后有关。
信号调节蛋白(SIRP)是一种穿模糖蛋白,属于跨膜蛋白家族。信号调节蛋白具有三个成员:SIRPα(CD172a)、SIRPβ(CD172b)、SIRPγ(CD172g)。这三个成员具有相似的胞外区域,但胞内结构域不同。胞外结构域包含三个免疫球蛋白(Ig)样区域,其中第一个Ig样区域为N端V样结构域(D1),第二、三个区域是两个C1样结构域。SIRPα(CD172a)的胞内结构域包含两个抑制性信号传导区域默契可以抑制信号转导以及相应的细胞功能。SIRP主要表达在巨噬细胞(M)、树突状细胞(DC)和神经元中。
CD47也是一种穿模糖蛋白,属于免疫球蛋白超家族,且在包括红细胞在内的所有细胞类型的表面表达,在细胞与细胞之间的通讯中具有多种功能。CD47与多种配体相互作用,例如整联蛋白、SIRPα、SIRPγ和血小板反应蛋白。CD47与SIRPα相互作用,CD47下调表达SIRPα的吞噬细胞的活性。研究发现,除正常组织表达CD47外,许多肿瘤细胞过度表达CD47,并以这种方式,传递“不要吃我”信号,从而避免被机体免疫监视发现而被先天免疫细胞杀死(免疫逃逸)。CD47抑制树突状细胞(DC)的成熟和激活。CD47还涉及细胞凋亡、存活、增殖、粘附、迁移和调节血管生成、血压、组织灌注或血小板稳态等过程。在癌症例如白血病、淋巴瘤、乳腺癌、结肠癌、卵巢癌、膀胱癌、前列腺癌和神经胶质瘤中,高水平的CD47与不良临床结果有关。迄今没有对于任何同时靶向MSLN和CD47的双特异性抗体。
发明内容
有鉴于此,本发明的目的在于提供一种重组双功能融合蛋白及应用,所述融合蛋白同时与MSLN以及CD47结合,从而发挥对MSLN阳性肿瘤细胞的治疗作用,并且阻断CD47与巨噬细胞表面上的SIRP结合,刺激巨噬细胞对肿瘤细胞的吞噬作用,具有显著的抗肿瘤活性。
为了实现上述发明目的,本发明提供以下技术方案:
本发明提供了一种重组双功能融合蛋白,所述融合蛋白由抗人MSLN抗体与结合人CD47蛋白的单域结构连接而成,所述重组双功能融合蛋白包括第一结合结构域和第二结合结构域;
所述第一结合结构域特异性结合靶分子人MSLN蛋白;所述第二结合结构域特异性结合靶分子CD47蛋白。
优选的,所述结合人CD47蛋白的单域结构包括衍生自人信号调节蛋白膜外N端V样结构域(D1)。
优选的,所述衍生自人信号调节蛋白膜外N端V样结构域(D1)的氨基酸序列如SEQID NO.1所示。
优选的,所述结合人CD47蛋白的单域结构还包括与SEQ ID NO.1所示序列具有至少80%、85%、90%、95%、98%或99%相似性的氨基酸。
优选的,所述融合蛋白包括重链和轻链,所述重链包括多肽链VH
优选的,所述肽链VH
优选的,所述融合蛋白包括重链和轻链,所述重链包括多肽链VH
优选的,所述多肽链VH
优选的,所述融合蛋白包括重链和轻链,所述重链包括多肽链V
优选的,所述肽链V
优选的,所述Fc的结构域来源包括野生型或突变型;连接子X为由多个相同或不同氨基酸组成的多肽链。
本发明还提供了一种治疗MSLN阳性肿瘤的药物,所述药物的有效成分包括上述融合蛋白。
本发明提供了一种重组双功能融合蛋白(MSLNmAb-V
附图说明
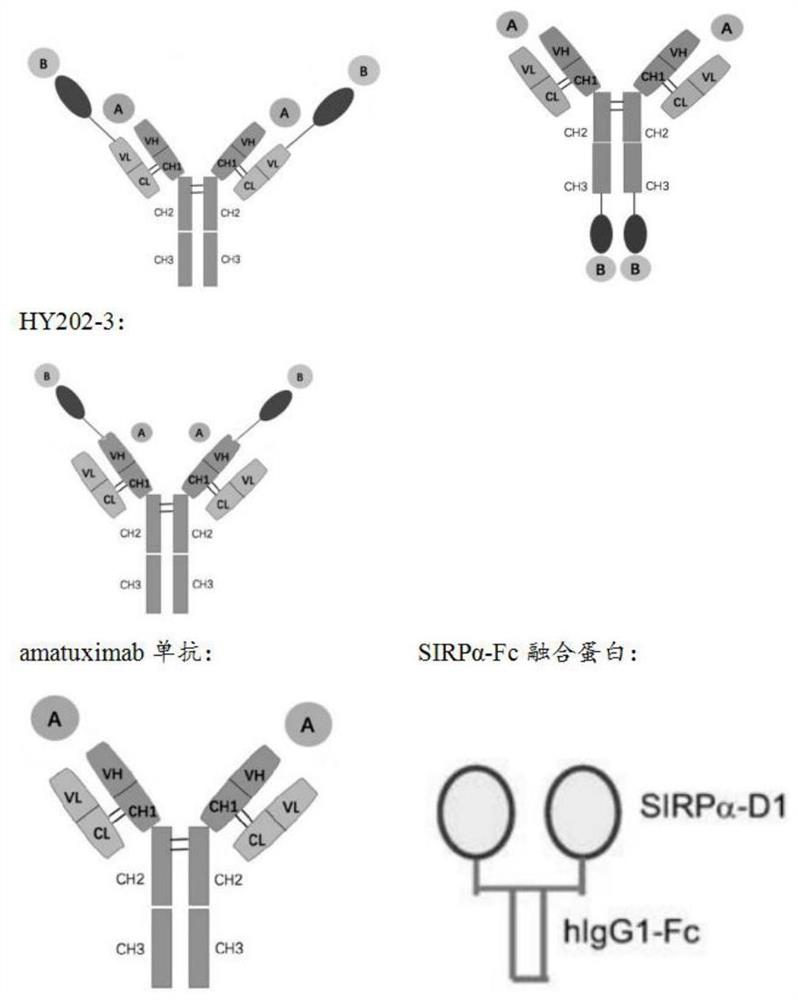
图1为融合蛋白结构的示意图;图1中A为MSLN抗原;B为CD47抗原;
图2为DOT检测蛋白表达;
图3靶点结合活性测定,其中图3A为靶点结合活性(MSLN),图3B为靶点结合活性(human CD47);
图4为HY202-1、HY202-2、HY202-3对ADCC效应细胞激活活性的影响,其中图4A为NCIH226报告基因测定,图4B为OVCAR-3报告基因测定;
图5为HY202-1、HY202-2、HY202-3对巨噬细胞吞噬活性的影响;
图6为HY202-1、HY202-2、HY202-3的体内抗肿瘤活性。
具体实施方式
本发明提供了一种重组双功能融合蛋白(MSLNmAb-SIRPα),所述融合蛋白由抗人MSLN抗体与结合人CD47蛋白的单域结构连接而成,所述重组双功能融合蛋白包括第一结合结构域和第二结合结构域;
所述第一结合结构域特异性结合靶分子人MSLN蛋白;所述第二结合结构域特异性结合靶分子CD47蛋白。
本发明所述结合人CD47蛋白的单域结构优选包括人信号调节蛋白(SIRPα)膜外N端V样结构域(D1),且所述人信号调节蛋白膜外N端V样结构域(D1)的氨基酸序列优选如SEQID NO.1所示。在本发明中,所述结合人CD47蛋白的单域结构优选还包括与SEQ ID NO.1所示序列具有至少80%、85%、90%、95%、98%或99%相似性的氨基酸。
本发明所述融化蛋白有三种不同的结构,具体结构示意图如图1所示,第一种结构的融合蛋白包括多肽链VH
本发明所述肽链VH
本发明所述Fc的结构域来源优选包括野生型或突变型,更优选为野生型,在本发明实施例中,采用的野生型Fc的氨基酸序列优选如SEQ ID NO.8所示。本发明所述连接子X优选为由多个相同或不同氨基酸组成的多肽链,在本发明实施例中所述连接单元X的氨基酸序列优选如SEQ ID NO.9所示。
本发明所述重组双功能融合蛋白中,人MSLN抗体可杀伤MSLN阳性表达的肿瘤细胞,SIRPα可激活巨噬细胞吞噬CD47阳性表达的肿瘤细胞,将两者结合后,获得上述三种不同的结构,可通过MSLN与CD47两种信号途径杀伤肿瘤细胞。
本发明对所述融合蛋白的制备方法并没有特殊限定,优选的包括人工合成编码各融合蛋白的多肽链的核苷酸序列,并分别克隆至载体,从而分别构建得到HY202-1、HY202-2和HY202-3表达载体;利用所述表达载体电转CHO-K1细胞,诱导蛋白表达并纯化。本发明对所述电转、诱导蛋白表达和纯化的方法并没有特殊限定,利用本领域的常规技术手段即可。
本发明还提供了上述融合蛋白在制备治疗MSLN阳性肿瘤的药物中的应用。本发明所述融合蛋白能够同时与MSLN以及CD47结合,从而通过抗体依赖、细胞介导的细胞毒及补体依赖的细胞毒活性而发挥对MSLN阳性肿瘤细胞的治疗作用,并且阻断CD47与巨噬细胞表面上的SIRP结合,刺激巨噬细胞对肿瘤细胞的吞噬作用,具有显著的抗肿瘤活性;并且其肿瘤抑制效果优于单一抗MSLN单抗治疗;可以用来治疗抗MSLN单抗无效或耐受的肿瘤患者。
本发明还提供了一种治疗MSLN阳性肿瘤的药物,所述药物的有效成分包括上述融合蛋白。
下面结合实施例对本发明提供的一种重组双功能融合蛋白及应用进行详细的说明,但是不能把它们理解为对本发明保护范围的限定。
实施例1
1、表达载体构建
抗体重链采用Amatuximab单抗VH
将抗人MSLN抗体重链VH
将V
2、蛋白表达及纯化
预先准备CHO细胞培养基(CD CHO Medium+8mM GlutaMAX;Gibco,10743-029;Gibco,35050-061)于T75方瓶,37℃预热;准备处于对数生长期的CHO-K1细胞,1×10
3、蛋白表达检测
用DOT blot检测蛋白表达。具体步骤是:根据工作量选择合适大小的硝酸纤维素膜(BBI Life Sciences,F619511-0005),标记每孔对应的位置和方向;取细胞上清原液,96孔膜每孔点样2μL,带标准品(0.01mg/mL,0.1mg/mL,1mg/mL,10mg/mL),晾干;将膜浸没在封闭液(2.5g脱脂奶粉+50mL PBS)中,置于摇床1h,转速60~90rpm;取一支羊抗人IgG-HRP抗体(Jakson Immuno Research,109-035-098)加入到50mL封闭液中,置于摇床1h,转速60~90rpm;用PBS洗膜3~5次,除去脱脂奶粉,置于摇床,转速60~90rpm,每次10min;每张96孔板大小的NC膜用1mL ECL化学发光液(Thermo,34577);使用化学发光成像仪拍照,曝光时间设定为自动曝光,用图像软件对图片进行灰度分析。结果如图2所示,以标准品为基准,单抗与双抗产品均表达,且产量近1g/L。
4、稳定细胞株筛选
基因转染的细胞,电转后24~48h,即用加压培养基对细胞进行稀释,加入到96孔细胞培养板中,每孔含有10000个细胞;细胞融合度达60%,取上清进行DOT检测;选择表达量高的逐步进行扩大培养,检测HPLC-titer,以及相关质量参数分析,最终选择3~5个高表达克隆。经筛选后,选择对数生长期的细胞经有限稀释法进行进一步筛选,将具有最高表达水平的细胞株挑选出来冻存备用。
5、蛋白生产及纯化
将稳定表达细胞株(3×10
6、靶点结合活性
用Elisa法检测靶点结合活性:
分析MSLN的靶点结合活性,具体步骤是:将人MSLN抗原溶解至包被缓冲液中,稀释至0.5μg/mL;然后加入到96孔ELISA检测板中,每孔100μL,然后将ELISA板放置于4℃度冰箱过夜。检测时,用洗涤液(TBS,0.05%Tween-20,pH 7.4)洗涤3次;用封闭液(洗涤液,2%BSA)先37℃封闭1.5h,经洗涤液洗涤3次;然后加入经过稀释的实验组抗体(设置Amatuximab单抗作阳性对照,IgG1阴性对照),37℃孵育1h,经洗涤液洗涤3次;然后加入辣根过氧化物酶(HRP)标记的、针对人IgG Fc片段的山羊抗体(Jakson Immuno Research,109-035-098),室温孵育1h;洗涤6次后加入HRP的底物,避光显色反应10~15min后加入终止液(1M H
表1靶点结合活性
用流式细胞术检测分析CD47的靶点结合活性,具体步骤是:取10
表2靶点结合活性
7、抗体依赖性细胞毒性
将对数生长期的10
Response(Relative luminescence units)=(处理样品数值-背景值)/(无抗体对照组-背景值);
利用GraphPad绘制曲线:纵坐标为光信号值,横坐标为药物浓度的对数值;据此可计算该抗体效应的EC50数值。结果如表3~4和图4所示,人肺鳞癌细胞NCIH226及乳腺癌细胞OVCAR-3中,双抗产品均明显激活ADCC功能,且效果优于单抗产品。
表3 NCIH226细胞报告基因测定
表4 OVCAR3报告基因测定
8、抗体依赖性细胞吞噬
将小鼠巨噬细胞(Raw264.7)加入到96孔细胞培养板中,每孔5×10
9、体内抗肿瘤药效实验
利用NCIH226细胞皮下肿瘤模型,对MSLNmAb-SIRPα的体内抗肿瘤活性进行研究。给30只裸鼠尾皮下注射NCIH226细胞,每只小鼠注射3×10
以上所述仅是本发明的优选实施方式,应当指出,对于本技术领域的普通技术人员来说,在不脱离本发明原理的前提下,还可以做出若干改进和润饰,这些改进和润饰也应视为本发明的保护范围。
序列表
<110> 南京华岩生物技术有限公司
河北神宇生物技术有限公司
<120> 一种重组双功能融合蛋白及应用
<141> 2020-11-30
<160> 16
<170> SIPOSequenceListing 1.0
<210> 1
<211> 133
<212> PRT
<213> 人工序列(Artificial Sequence)
<400> 1
Glu Glu Glu Leu Gln Val Ile Gln Pro Asp Lys Ser Val Ser Val Ala
1 5 10 15
Ala Gly Glu Ser Ala Ile Leu His Cys Thr Val Thr Ser Leu Ile Pro
20 25 30
Val Gly Pro Ile Gln Trp Phe Arg Gly Ala Gly Pro Ala Arg Glu Leu
35 40 45
Ile Tyr Asn Gln Lys Glu Gly His Phe Pro Arg Val Thr Thr Val Ser
50 55 60
Glu Ser Thr Lys Arg Glu Asn Met Asp Phe Ser Ile Ser Ile Ser Ala
65 70 75 80
Ile Thr Pro Ala Asp Ala Gly Thr Tyr Tyr Cys Val Lys Phe Arg Lys
85 90 95
Gly Ser Pro Asp Thr Glu Phe Lys Ser Gly Ala Gly Thr Glu Leu Ser
100 105 110
Val Arg Ala Lys Pro Ser Ala Pro Val Val Ser Gly Pro Ala Ala Arg
115 120 125
Ala Thr Pro Gln His
130
<210> 2
<211> 449
<212> PRT
<213> 人工序列(Artificial Sequence)
<400> 2
Gln Val Gln Leu Gln Gln Ser Gly Pro Glu Leu Glu Lys Pro Gly Ala
1 5 10 15
Ser Val Lys Ile Ser Cys Lys Ala Ser Gly Tyr Ser Phe Thr Gly Tyr
20 25 30
Thr Met Asn Trp Val Lys Gln Ser His Gly Lys Ser Leu Glu Trp Ile
35 40 45
Gly Leu Ile Thr Pro Tyr Asn Gly Ala Ser Ser Tyr Asn Gln Lys Phe
50 55 60
Arg Gly Lys Ala Thr Leu Thr Val Asp Lys Ser Ser Ser Thr Ala Tyr
65 70 75 80
Met Asp Leu Leu Ser Leu Thr Ser Glu Asp Ser Ala Val Tyr Phe Cys
85 90 95
Ala Arg Gly Gly Tyr Asp Gly Arg Gly Phe Asp Tyr Trp Gly Ser Gly
100 105 110
Thr Pro Val Thr Val Ser Ser Ala Ser Thr Lys Gly Pro Ser Val Phe
115 120 125
Pro Leu Ala Pro Ser Ser Lys Ser Thr Ser Gly Gly Thr Ala Ala Leu
130 135 140
Gly Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro Val Thr Val Ser Trp
145 150 155 160
Asn Ser Gly Ala Leu Thr Ser Gly Val His Thr Phe Pro Ala Val Leu
165 170 175
Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val Val Thr Val Pro Ser
180 185 190
Ser Ser Leu Gly Thr Gln Thr Tyr Ile Cys Asn Val Asn His Lys Pro
195 200 205
Ser Asn Thr Lys Val Asp Lys Lys Val Glu Pro Lys Ser Cys Asp Lys
210 215 220
Thr His Thr Cys Pro Pro Cys Pro Ala Pro Glu Leu Leu Gly Gly Pro
225 230 235 240
Ser Val Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met Ile Ser
245 250 255
Arg Thr Pro Glu Val Thr Cys Val Val Val Asp Val Ser His Glu Asp
260 265 270
Pro Glu Val Lys Phe Asn Trp Tyr Val Asp Gly Val Glu Val His Asn
275 280 285
Ala Lys Thr Lys Pro Arg Glu Glu Gln Tyr Asn Ser Thr Tyr Arg Val
290 295 300
Val Ser Val Leu Thr Val Leu His Gln Asp Trp Leu Asn Gly Lys Glu
305 310 315 320
Tyr Lys Cys Lys Val Ser Asn Lys Ala Leu Pro Ala Pro Ile Glu Lys
325 330 335
Thr Ile Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro Gln Val Tyr Thr
340 345 350
Leu Pro Pro Ser Arg Asp Glu Leu Thr Lys Asn Gln Val Ser Leu Thr
355 360 365
Cys Leu Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp Glu
370 375 380
Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro Val Leu
385 390 395 400
Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val Asp Lys
405 410 415
Ser Arg Trp Gln Gln Gly Asn Val Phe Ser Cys Ser Val Met His Glu
420 425 430
Ala Leu His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser Pro Gly
435 440 445
Lys
<210> 3
<211> 361
<212> PRT
<213> 人工序列(Artificial Sequence)
<400> 3
Glu Glu Glu Leu Gln Val Ile Gln Pro Asp Lys Ser Val Ser Val Ala
1 5 10 15
Ala Gly Glu Ser Ala Ile Leu His Cys Thr Val Thr Ser Leu Ile Pro
20 25 30
Val Gly Pro Ile Gln Trp Phe Arg Gly Ala Gly Pro Ala Arg Glu Leu
35 40 45
Ile Tyr Asn Gln Lys Glu Gly His Phe Pro Arg Val Thr Thr Val Ser
50 55 60
Glu Ser Thr Lys Arg Glu Asn Met Asp Phe Ser Ile Ser Ile Ser Ala
65 70 75 80
Ile Thr Pro Ala Asp Ala Gly Thr Tyr Tyr Cys Val Lys Phe Arg Lys
85 90 95
Gly Ser Pro Asp Thr Glu Phe Lys Ser Gly Ala Gly Thr Glu Leu Ser
100 105 110
Val Arg Ala Lys Pro Ser Ala Pro Val Val Ser Gly Pro Ala Ala Arg
115 120 125
Ala Thr Pro Gln His Gly Gly Gly Gly Ser Gly Gly Gly Gly Ser Gly
130 135 140
Gly Gly Gly Ser Asp Ile Glu Leu Thr Gln Ser Pro Ala Ile Met Ser
145 150 155 160
Ala Ser Pro Gly Glu Lys Val Thr Met Thr Cys Ser Ala Ser Ser Ser
165 170 175
Val Ser Tyr Met His Trp Tyr Gln Gln Lys Ser Gly Thr Ser Pro Lys
180 185 190
Arg Trp Ile Tyr Asp Thr Ser Lys Leu Ala Ser Gly Val Pro Gly Arg
195 200 205
Phe Ser Gly Ser Gly Ser Gly Asn Ser Tyr Ser Leu Thr Ile Ser Ser
210 215 220
Val Glu Ala Glu Asp Asp Ala Thr Tyr Tyr Cys Gln Gln Trp Ser Lys
225 230 235 240
His Pro Leu Thr Phe Gly Ser Gly Thr Lys Val Glu Ile Lys Arg Thr
245 250 255
Val Ala Ala Pro Ser Val Phe Ile Phe Pro Pro Ser Asp Glu Gln Leu
260 265 270
Lys Ser Gly Thr Ala Ser Val Val Cys Leu Leu Asn Asn Phe Tyr Pro
275 280 285
Arg Glu Ala Lys Val Gln Trp Lys Val Asp Asn Ala Leu Gln Ser Gly
290 295 300
Asn Ser Gln Glu Ser Val Thr Glu Gln Asp Ser Lys Asp Ser Thr Tyr
305 310 315 320
Ser Leu Ser Ser Thr Leu Thr Leu Ser Lys Ala Asp Tyr Glu Lys His
325 330 335
Lys Val Tyr Ala Cys Glu Val Thr His Gln Gly Leu Ser Ser Pro Val
340 345 350
Thr Lys Ser Phe Asn Arg Gly Glu Cys
355 360
<210> 4
<211> 597
<212> PRT
<213> 人工序列(Artificial Sequence)
<400> 4
Gln Val Gln Leu Gln Gln Ser Gly Pro Glu Leu Glu Lys Pro Gly Ala
1 5 10 15
Ser Val Lys Ile Ser Cys Lys Ala Ser Gly Tyr Ser Phe Thr Gly Tyr
20 25 30
Thr Met Asn Trp Val Lys Gln Ser His Gly Lys Ser Leu Glu Trp Ile
35 40 45
Gly Leu Ile Thr Pro Tyr Asn Gly Ala Ser Ser Tyr Asn Gln Lys Phe
50 55 60
Arg Gly Lys Ala Thr Leu Thr Val Asp Lys Ser Ser Ser Thr Ala Tyr
65 70 75 80
Met Asp Leu Leu Ser Leu Thr Ser Glu Asp Ser Ala Val Tyr Phe Cys
85 90 95
Ala Arg Gly Gly Tyr Asp Gly Arg Gly Phe Asp Tyr Trp Gly Ser Gly
100 105 110
Thr Pro Val Thr Val Ser Ser Ala Ser Thr Lys Gly Pro Ser Val Phe
115 120 125
Pro Leu Ala Pro Ser Ser Lys Ser Thr Ser Gly Gly Thr Ala Ala Leu
130 135 140
Gly Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro Val Thr Val Ser Trp
145 150 155 160
Asn Ser Gly Ala Leu Thr Ser Gly Val His Thr Phe Pro Ala Val Leu
165 170 175
Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val Val Thr Val Pro Ser
180 185 190
Ser Ser Leu Gly Thr Gln Thr Tyr Ile Cys Asn Val Asn His Lys Pro
195 200 205
Ser Asn Thr Lys Val Asp Lys Lys Val Glu Pro Lys Ser Cys Asp Lys
210 215 220
Thr His Thr Cys Pro Pro Cys Pro Ala Pro Glu Leu Leu Gly Gly Pro
225 230 235 240
Ser Val Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met Ile Ser
245 250 255
Arg Thr Pro Glu Val Thr Cys Val Val Val Asp Val Ser His Glu Asp
260 265 270
Pro Glu Val Lys Phe Asn Trp Tyr Val Asp Gly Val Glu Val His Asn
275 280 285
Ala Lys Thr Lys Pro Arg Glu Glu Gln Tyr Asn Ser Thr Tyr Arg Val
290 295 300
Val Ser Val Leu Thr Val Leu His Gln Asp Trp Leu Asn Gly Lys Glu
305 310 315 320
Tyr Lys Cys Lys Val Ser Asn Lys Ala Leu Pro Ala Pro Ile Glu Lys
325 330 335
Thr Ile Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro Gln Val Tyr Thr
340 345 350
Leu Pro Pro Ser Arg Asp Glu Leu Thr Lys Asn Gln Val Ser Leu Thr
355 360 365
Cys Leu Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp Glu
370 375 380
Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro Val Leu
385 390 395 400
Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val Asp Lys
405 410 415
Ser Arg Trp Gln Gln Gly Asn Val Phe Ser Cys Ser Val Met His Glu
420 425 430
Ala Leu His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser Pro Gly
435 440 445
Lys Gly Gly Gly Gly Ser Gly Gly Gly Gly Ser Gly Gly Gly Gly Ser
450 455 460
Glu Glu Glu Leu Gln Val Ile Gln Pro Asp Lys Ser Val Ser Val Ala
465 470 475 480
Ala Gly Glu Ser Ala Ile Leu His Cys Thr Val Thr Ser Leu Ile Pro
485 490 495
Val Gly Pro Ile Gln Trp Phe Arg Gly Ala Gly Pro Ala Arg Glu Leu
500 505 510
Ile Tyr Asn Gln Lys Glu Gly His Phe Pro Arg Val Thr Thr Val Ser
515 520 525
Glu Ser Thr Lys Arg Glu Asn Met Asp Phe Ser Ile Ser Ile Ser Ala
530 535 540
Ile Thr Pro Ala Asp Ala Gly Thr Tyr Tyr Cys Val Lys Phe Arg Lys
545 550 555 560
Gly Ser Pro Asp Thr Glu Phe Lys Ser Gly Ala Gly Thr Glu Leu Ser
565 570 575
Val Arg Ala Lys Pro Ser Ala Pro Val Val Ser Gly Pro Ala Ala Arg
580 585 590
Ala Thr Pro Gln His
595
<210> 5
<211> 213
<212> PRT
<213> 人工序列(Artificial Sequence)
<400> 5
Asp Ile Glu Leu Thr Gln Ser Pro Ala Ile Met Ser Ala Ser Pro Gly
1 5 10 15
Glu Lys Val Thr Met Thr Cys Ser Ala Ser Ser Ser Val Ser Tyr Met
20 25 30
His Trp Tyr Gln Gln Lys Ser Gly Thr Ser Pro Lys Arg Trp Ile Tyr
35 40 45
Asp Thr Ser Lys Leu Ala Ser Gly Val Pro Gly Arg Phe Ser Gly Ser
50 55 60
Gly Ser Gly Asn Ser Tyr Ser Leu Thr Ile Ser Ser Val Glu Ala Glu
65 70 75 80
Asp Asp Ala Thr Tyr Tyr Cys Gln Gln Trp Ser Lys His Pro Leu Thr
85 90 95
Phe Gly Ser Gly Thr Lys Val Glu Ile Lys Arg Thr Val Ala Ala Pro
100 105 110
Ser Val Phe Ile Phe Pro Pro Ser Asp Glu Gln Leu Lys Ser Gly Thr
115 120 125
Ala Ser Val Val Cys Leu Leu Asn Asn Phe Tyr Pro Arg Glu Ala Lys
130 135 140
Val Gln Trp Lys Val Asp Asn Ala Leu Gln Ser Gly Asn Ser Gln Glu
145 150 155 160
Ser Val Thr Glu Gln Asp Ser Lys Asp Ser Thr Tyr Ser Leu Ser Ser
165 170 175
Thr Leu Thr Leu Ser Lys Ala Asp Tyr Glu Lys His Lys Val Tyr Ala
180 185 190
Cys Glu Val Thr His Gln Gly Leu Ser Ser Pro Val Thr Lys Ser Phe
195 200 205
Asn Arg Gly Glu Cys
210
<210> 6
<211> 597
<212> PRT
<213> 人工序列(Artificial Sequence)
<400> 6
Glu Glu Glu Leu Gln Val Ile Gln Pro Asp Lys Ser Val Ser Val Ala
1 5 10 15
Ala Gly Glu Ser Ala Ile Leu His Cys Thr Val Thr Ser Leu Ile Pro
20 25 30
Val Gly Pro Ile Gln Trp Phe Arg Gly Ala Gly Pro Ala Arg Glu Leu
35 40 45
Ile Tyr Asn Gln Lys Glu Gly His Phe Pro Arg Val Thr Thr Val Ser
50 55 60
Glu Ser Thr Lys Arg Glu Asn Met Asp Phe Ser Ile Ser Ile Ser Ala
65 70 75 80
Ile Thr Pro Ala Asp Ala Gly Thr Tyr Tyr Cys Val Lys Phe Arg Lys
85 90 95
Gly Ser Pro Asp Thr Glu Phe Lys Ser Gly Ala Gly Thr Glu Leu Ser
100 105 110
Val Arg Ala Lys Pro Ser Ala Pro Val Val Ser Gly Pro Ala Ala Arg
115 120 125
Ala Thr Pro Gln His Gly Gly Gly Gly Ser Gly Gly Gly Gly Ser Gly
130 135 140
Gly Gly Gly Ser Gln Val Gln Leu Gln Gln Ser Gly Pro Glu Leu Glu
145 150 155 160
Lys Pro Gly Ala Ser Val Lys Ile Ser Cys Lys Ala Ser Gly Tyr Ser
165 170 175
Phe Thr Gly Tyr Thr Met Asn Trp Val Lys Gln Ser His Gly Lys Ser
180 185 190
Leu Glu Trp Ile Gly Leu Ile Thr Pro Tyr Asn Gly Ala Ser Ser Tyr
195 200 205
Asn Gln Lys Phe Arg Gly Lys Ala Thr Leu Thr Val Asp Lys Ser Ser
210 215 220
Ser Thr Ala Tyr Met Asp Leu Leu Ser Leu Thr Ser Glu Asp Ser Ala
225 230 235 240
Val Tyr Phe Cys Ala Arg Gly Gly Tyr Asp Gly Arg Gly Phe Asp Tyr
245 250 255
Trp Gly Ser Gly Thr Pro Val Thr Val Ser Ser Ala Ser Thr Lys Gly
260 265 270
Pro Ser Val Phe Pro Leu Ala Pro Ser Ser Lys Ser Thr Ser Gly Gly
275 280 285
Thr Ala Ala Leu Gly Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro Val
290 295 300
Thr Val Ser Trp Asn Ser Gly Ala Leu Thr Ser Gly Val His Thr Phe
305 310 315 320
Pro Ala Val Leu Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val Val
325 330 335
Thr Val Pro Ser Ser Ser Leu Gly Thr Gln Thr Tyr Ile Cys Asn Val
340 345 350
Asn His Lys Pro Ser Asn Thr Lys Val Asp Lys Lys Val Glu Pro Lys
355 360 365
Ser Cys Asp Lys Thr His Thr Cys Pro Pro Cys Pro Ala Pro Glu Leu
370 375 380
Leu Gly Gly Pro Ser Val Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr
385 390 395 400
Leu Met Ile Ser Arg Thr Pro Glu Val Thr Cys Val Val Val Asp Val
405 410 415
Ser His Glu Asp Pro Glu Val Lys Phe Asn Trp Tyr Val Asp Gly Val
420 425 430
Glu Val His Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln Tyr Asn Ser
435 440 445
Thr Tyr Arg Val Val Ser Val Leu Thr Val Leu His Gln Asp Trp Leu
450 455 460
Asn Gly Lys Glu Tyr Lys Cys Lys Val Ser Asn Lys Ala Leu Pro Ala
465 470 475 480
Pro Ile Glu Lys Thr Ile Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro
485 490 495
Gln Val Tyr Thr Leu Pro Pro Ser Arg Asp Glu Leu Thr Lys Asn Gln
500 505 510
Val Ser Leu Thr Cys Leu Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala
515 520 525
Val Glu Trp Glu Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr
530 535 540
Pro Pro Val Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu
545 550 555 560
Thr Val Asp Lys Ser Arg Trp Gln Gln Gly Asn Val Phe Ser Cys Ser
565 570 575
Val Met His Glu Ala Leu His Asn His Tyr Thr Gln Lys Ser Leu Ser
580 585 590
Leu Ser Pro Gly Lys
595
<210> 7
<211> 213
<212> PRT
<213> 人工序列(Artificial Sequence)
<400> 7
Asp Ile Glu Leu Thr Gln Ser Pro Ala Ile Met Ser Ala Ser Pro Gly
1 5 10 15
Glu Lys Val Thr Met Thr Cys Ser Ala Ser Ser Ser Val Ser Tyr Met
20 25 30
His Trp Tyr Gln Gln Lys Ser Gly Thr Ser Pro Lys Arg Trp Ile Tyr
35 40 45
Asp Thr Ser Lys Leu Ala Ser Gly Val Pro Gly Arg Phe Ser Gly Ser
50 55 60
Gly Ser Gly Asn Ser Tyr Ser Leu Thr Ile Ser Ser Val Glu Ala Glu
65 70 75 80
Asp Asp Ala Thr Tyr Tyr Cys Gln Gln Trp Ser Lys His Pro Leu Thr
85 90 95
Phe Gly Ser Gly Thr Lys Val Glu Ile Lys Arg Thr Val Ala Ala Pro
100 105 110
Ser Val Phe Ile Phe Pro Pro Ser Asp Glu Gln Leu Lys Ser Gly Thr
115 120 125
Ala Ser Val Val Cys Leu Leu Asn Asn Phe Tyr Pro Arg Glu Ala Lys
130 135 140
Val Gln Trp Lys Val Asp Asn Ala Leu Gln Ser Gly Asn Ser Gln Glu
145 150 155 160
Ser Val Thr Glu Gln Asp Ser Lys Asp Ser Thr Tyr Ser Leu Ser Ser
165 170 175
Thr Leu Thr Leu Ser Lys Ala Asp Tyr Glu Lys His Lys Val Tyr Ala
180 185 190
Cys Glu Val Thr His Gln Gly Leu Ser Ser Pro Val Thr Lys Ser Phe
195 200 205
Asn Arg Gly Glu Cys
210
<210> 8
<211> 330
<212> PRT
<213> 人工序列(Artificial Sequence)
<400> 8
Ala Ser Thr Lys Gly Pro Ser Val Phe Pro Leu Ala Pro Ser Ser Lys
1 5 10 15
Ser Thr Ser Gly Gly Thr Ala Ala Leu Gly Cys Leu Val Lys Asp Tyr
20 25 30
Phe Pro Glu Pro Val Thr Val Ser Trp Asn Ser Gly Ala Leu Thr Ser
35 40 45
Gly Val His Thr Phe Pro Ala Val Leu Gln Ser Ser Gly Leu Tyr Ser
50 55 60
Leu Ser Ser Val Val Thr Val Pro Ser Ser Ser Leu Gly Thr Gln Thr
65 70 75 80
Tyr Ile Cys Asn Val Asn His Lys Pro Ser Asn Thr Lys Val Asp Lys
85 90 95
Lys Val Glu Pro Lys Ser Cys Asp Lys Thr His Thr Cys Pro Pro Cys
100 105 110
Pro Ala Pro Glu Leu Leu Gly Gly Pro Ser Val Phe Leu Phe Pro Pro
115 120 125
Lys Pro Lys Asp Thr Leu Met Ile Ser Arg Thr Pro Glu Val Thr Cys
130 135 140
Val Val Val Asp Val Ser His Glu Asp Pro Glu Val Lys Phe Asn Trp
145 150 155 160
Tyr Val Asp Gly Val Glu Val His Asn Ala Lys Thr Lys Pro Arg Glu
165 170 175
Glu Gln Tyr Asn Ser Thr Tyr Arg Val Val Ser Val Leu Thr Val Leu
180 185 190
His Gln Asp Trp Leu Asn Gly Lys Glu Tyr Lys Cys Lys Val Ser Asn
195 200 205
Lys Ala Leu Pro Ala Pro Ile Glu Lys Thr Ile Ser Lys Ala Lys Gly
210 215 220
Gln Pro Arg Glu Pro Gln Val Tyr Thr Leu Pro Pro Ser Arg Asp Glu
225 230 235 240
Leu Thr Lys Asn Gln Val Ser Leu Thr Cys Leu Val Lys Gly Phe Tyr
245 250 255
Pro Ser Asp Ile Ala Val Glu Trp Glu Ser Asn Gly Gln Pro Glu Asn
260 265 270
Asn Tyr Lys Thr Thr Pro Pro Val Leu Asp Ser Asp Gly Ser Phe Phe
275 280 285
Leu Tyr Ser Lys Leu Thr Val Asp Lys Ser Arg Trp Gln Gln Gly Asn
290 295 300
Val Phe Ser Cys Ser Val Met His Glu Ala Leu His Asn His Tyr Thr
305 310 315 320
Gln Lys Ser Leu Ser Leu Ser Pro Gly Lys
325 330
<210> 9
<211> 15
<212> PRT
<213> 人工序列(Artificial Sequence)
<400> 9
Gly Gly Gly Gly Ser Gly Gly Gly Gly Ser Gly Gly Gly Gly Ser
1 5 10 15
<210> 10
<211> 1347
<212> DNA
<213> 人工序列(Artificial Sequence)
<400> 10
caggtgcagc tgcagcagtc tggaccagag ctggagaagc ctggagcctc tgtgaagatc 60
tcctgtaagg cttctggcta ctccttcacc ggctatacaa tgaactgggt gaagcagagc 120
catggcaagt ctctggagtg gatcggcctg atcacccctt acaacggcgc ctccagctat 180
aatcagaagt ttaggggcaa ggctaccctg acagtggaca agtcttccag caccgcctat 240
atggacctgc tgagcctgac atctgaggat tccgccgtgt acttctgcgc taggggcgga 300
tatgacggaa ggggctttga ttactggggc tccggcaccc ctgtgacagt gtcttccgct 360
tccaccaagg gcccaagcgt gtttccactg gcccccagct ctaagagcac ctctggagga 420
acagccgctc tgggctgtct ggtgaaggat tacttcccag agcccgtgac agtgagctgg 480
aactctggcg ccctgacctc cggagtgcac acatttcccg ctgtgctgca gtccagcggc 540
ctgtatagcc tgtcttccgt ggtgaccgtg cctagctctt ccctgggcac ccagacatac 600
atctgcaacg tgaatcacaa gccctccaat acaaaggtgg acaagaaggt ggagcctaag 660
agctgtgata agacccatac atgcccccct tgtcctgctc cagagctgct gggcggacca 720
tccgtgttcc tgtttccacc caagcccaag gacaccctga tgatctccag aacccctgag 780
gtgacatgcg tggtggtgga cgtgtcccac gaggatccag aggtgaagtt caactggtac 840
gtggatggcg tggaggtgca taatgccaag accaagccaa gagaggagca gtacaattct 900
acctatcgcg tggtgtccgt gctgacagtg ctgcaccagg actggctgaa cggcaaggag 960
tacaagtgca aggtgagcaa taaggccctg cccgctccta tcgagaagac catctctaag 1020
gctaagggcc agcccaggga gccacaggtg tacaccctgc ctccaagccg ggacgagctg 1080
accaagaacc aggtgtctct gacatgtctg gtgaagggct tctacccatc tgacatcgcc 1140
gtggagtggg agtccaatgg ccagcccgag aacaattata agaccacacc ccctgtgctg 1200
gactccgatg gcagcttctt tctgtactcc aagctgaccg tggataagag ccgctggcag 1260
cagggcaacg tgttttcctg tagcgtgatg catgaggctc tgcacaatca ttacacacag 1320
aagtctctgt ccctgagccc cggcaag 1347
<210> 11
<211> 399
<212> DNA
<213> 人工序列(Artificial Sequence)
<400> 11
gaggaggagc tgcaggtcat ccagccagac aagtccgtga gcgtggctgc tggagagagc 60
gccatcctgc attgtaccgt gacatctctg atcccagtgg gaccaatcca gtggtttagg 120
ggagctggac ctgctcggga gctgatctac aaccagaagg agggccactt cccaagagtg 180
accacagtgt ctgagtccac caagcgcgag aatatggact ttagcatctc tatctccgct 240
atcaccccag ccgatgctgg cacatactat tgcgtgaagt tcagaaaggg ctcccccgat 300
accgagttta agagcggagc tggaacagag ctgtctgtga gggccaagcc ttctgctcca 360
gtggtgtccg gccccgccgc tcgggctacc cctcagcac 399
<210> 12
<211> 639
<212> DNA
<213> 人工序列(Artificial Sequence)
<400> 12
gacatcgagc tgacccagag cccagctatc atgtccgcct ccccaggaga gaaggtgacc 60
atgacatgtt ccgcctcctc ctccgtgtcc tacatgcatt ggtatcagca gaagtctggc 120
acctccccca agagatggat ctacgataca agcaagctgg cttctggagt gcctggccgc 180
ttctccggaa gcggatctgg caactcctat agcctgacca tctccagcgt ggaggccgag 240
gacgatgcta catactattg ccagcagtgg tctaagcatc ctctgacctt tggctccggc 300
acaaaggtgg agatcaagag gaccgtggcc gctccatccg tgttcatctt tccccctagc 360
gacgagcagc tgaagtccgg cacagccagc gtggtgtgcc tgctgaacaa tttctacccc 420
cgggaggcca aggtgcagtg gaaggtggat aacgctctgc agagcggcaa ttctcaggag 480
tccgtgaccg agcaggacag caaggattct acatattccc tgtcttccac cctgacactg 540
tctaaggccg actacgagaa gcacaaggtg tatgcttgcg aggtgaccca tcagggcctg 600
agctctcctg tgacaaagtc ctttaatcgc ggcgagtgt 639
<210> 13
<211> 1083
<212> DNA
<213> 人工序列(Artificial Sequence)
<400> 13
gaggaggagc tgcaggtcat ccagccagac aagtccgtga gcgtggctgc tggagagagc 60
gccatcctgc attgtaccgt gacatctctg atcccagtgg gaccaatcca gtggtttagg 120
ggagctggac ctgctcggga gctgatctac aaccagaagg agggccactt cccaagagtg 180
accacagtgt ctgagtccac caagcgcgag aatatggact ttagcatctc tatctccgct 240
atcaccccag ccgatgctgg cacatactat tgcgtgaagt tcagaaaggg ctcccccgat 300
accgagttta agagcggagc tggaacagag ctgtctgtga gggccaagcc ttctgctcca 360
gtggtgtccg gccccgccgc tcgggctacc cctcagcacg gaggaggagg aagcggcgga 420
ggaggctctg gcggcggcgg ctccgacatc gagctgaccc agagcccagc tatcatgtcc 480
gcctccccag gagagaaggt gaccatgaca tgttccgcct cctcctccgt gtcctacatg 540
cattggtatc agcagaagtc tggcacctcc cccaagagat ggatctacga tacaagcaag 600
ctggcttctg gagtgcctgg ccgcttctcc ggaagcggat ctggcaactc ctatagcctg 660
accatctcca gcgtggaggc cgaggacgat gctacatact attgccagca gtggtctaag 720
catcctctga cctttggctc cggcacaaag gtggagatca agaggaccgt ggccgctcca 780
tccgtgttca tctttccccc tagcgacgag cagctgaagt ccggcacagc cagcgtggtg 840
tgcctgctga acaatttcta cccccgggag gccaaggtgc agtggaaggt ggataacgct 900
ctgcagagcg gcaattctca ggagtccgtg accgagcagg acagcaagga ttctacatat 960
tccctgtctt ccaccctgac actgtctaag gccgactacg agaagcacaa ggtgtatgct 1020
tgcgaggtga cccatcaggg cctgagctct cctgtgacaa agtcctttaa tcgcggcgag 1080
tgt 1083
<210> 14
<211> 1791
<212> DNA
<213> 人工序列(Artificial Sequence)
<400> 14
caggtgcagc tgcagcagtc tggacctgag ctggagaagc caggagcctc cgtgaagatc 60
agctgtaagg cttccggcta cagcttcacc ggctatacaa tgaactgggt gaagcagtct 120
catggcaagt ccctggagtg gatcggcctg atcaccccat acaacggcgc ctccagctat 180
aatcagaagt ttagaggcaa ggctaccctg acagtggaca agtcttccag caccgcctat 240
atggacctgc tgtctctgac atccgaggat agcgccgtgt acttctgcgc taggggcgga 300
tatgacggaa ggggctttga ttactggggc tctggaaccc ctgtgaccgt gtcctccgcc 360
tccaccaagg gaccatccgt gttcccactg gcccccagct ctaagagcac ctctggagga 420
acagccgctc tgggctgtct ggtgaaggat tacttcccag agcccgtgac agtgtcttgg 480
aactccggcg ccctgacctc tggagtgcac acatttcctg ctgtgctgca gtccagcggc 540
ctgtattccc tgtcttccgt ggtgaccgtg ccaagctctt ccctgggcac ccagacatac 600
atctgcaacg tgaatcacaa gccctctaat acaaaggtgg acaagaaggt ggagcctaag 660
tcctgtgata agacccatac atgcccccct tgtcctgctc cagagctgct gggcggacca 720
agcgtgttcc tgtttccacc caagcccaag gacaccctga tgatcagccg cacccctgag 780
gtgacatgcg tggtggtgga cgtgtctcac gaggatccag aggtgaagtt caactggtac 840
gtggatggcg tggaggtgca taatgccaag accaagccca gggaggagca gtacaattct 900
acctatcggg tggtgtccgt gctgacagtg ctgcaccagg actggctgaa cggcaaggag 960
tacaagtgca aggtgtccaa taaggccctg cccgctccta tcgagaagac catcagcaag 1020
gctaagggcc agcccaggga gccacaggtg tacaccctgc ctccatcccg ggacgagctg 1080
accaagaacc aggtgagcct gacatgtctg gtgaagggct tctacccatc tgacatcgcc 1140
gtggagtggg agtccaatgg ccagcccgag aacaattata agaccacacc ccctgtgctg 1200
gacagcgatg gctctttctt tctgtactcc aagctgaccg tggacaagag caggtggcag 1260
cagggcaacg tgttttcctg cagcgtgatg catgaggctc tgcacaatca ttatacacag 1320
aagtctctgt ccctgagccc cggcaaggga ggaggaggat ccggaggagg aggaagcggc 1380
ggcggcggct ctgaggagga gctgcaggtc atccagcctg ataagtccgt gtccgtggct 1440
gctggagaga gcgccatcct gcactgtacc gtgacatctc tgatccccgt gggccctatc 1500
cagtggttta gaggagctgg accagctcgc gagctgatct acaaccagaa ggagggccat 1560
ttccctagag tgaccacagt gagcgagtct accaagcgcg agaatatgga cttttccatc 1620
agcatctctg ccatcacccc agccgatgct ggcacatact attgcgtgaa gttccggaag 1680
ggctctcccg ataccgagtt taagtccggc gctggcacag agctgagcgt gagagccaag 1740
ccatccgctc ccgtggtgag cggccccgcc gctcgcgcca cccctcagca t 1791
<210> 15
<211> 639
<212> DNA
<213> 人工序列(Artificial Sequence)
<400> 15
gacatcgagc tgacccagag cccagctatc atgtccgcct ccccaggaga gaaggtgacc 60
atgacatgtt ccgcctcctc ctccgtgtcc tacatgcatt ggtatcagca gaagagcggc 120
acctctccta agaggtggat ctacgataca tccaagctgg cctccggagt gccaggccgg 180
ttctctggat ccggaagcgg caactcttat tccctgacca tctccagcgt ggaggccgag 240
gacgatgcta catactattg ccagcagtgg agcaagcacc ccctgacctt tggctctggc 300
acaaaggtgg agatcaagag aaccgtggcc gctccttccg tgttcatctt tcccccttcc 360
gacgagcagc tgaagtctgg cacagcctcc gtggtgtgcc tgctgaacaa tttctaccca 420
cgcgaggcca aggtgcagtg gaaggtggat aacgctctgc agtccggcaa tagccaggag 480
tctgtgaccg agcaggactc caaggatagc acatattctc tgtcttccac cctgacactg 540
tccaaggccg actacgagaa gcacaaggtg tatgcttgcg aggtgaccca tcagggcctg 600
agctctcccg tgacaaagag ctttaatagg ggcgagtgt 639
<210> 16
<211> 1791
<212> DNA
<213> 人工序列(Artificial Sequence)
<400> 16
gaggaggagc tgcaggtcat ccagccagac aagagcgtgt ctgtggccgc tggcgagtct 60
gctatcctgc attgtaccgt gacatccctg atcccagtgg gaccaatcca gtggtttagg 120
ggagctggac ctgctaggga gctgatctat aaccagaagg agggccactt cccaagagtg 180
accacagtgt ccgagagcac caagcgcgag aatatggact tttctatctc catcagcgcc 240
atcaccccag ccgatgctgg cacatactat tgcgtgaagt tcaggaaggg cagccccgat 300
accgagttta agtctggcgc tggcacagag ctgtccgtga gagccaagcc ttccgcccca 360
gtggtgtctg gccccgccgc tcgcgccacc cctcagcatg gaggaggagg atccggagga 420
ggaggcagcg gcggcggcgg ctctcaggtg cagctgcagc agagcggacc tgagctggag 480
aagccaggag cctccgtgaa gatcagctgt aaggcttccg gctacagctt caccggctat 540
acaatgaact gggtgaagca gtctcatggc aagtccctgg agtggatcgg cctgatcacc 600
ccttacaacg gcgcctccag ctataatcag aagtttcggg gcaaggctac cctgacagtg 660
gacaagtctt ccagcaccgc ctatatggac ctgctgtctc tgacatccga ggatagcgcc 720
gtgtacttct gcgctagagg aggatatgac ggaaggggct ttgattactg gggctctggc 780
acccctgtga cagtgtcttc cgcttccacc aagggaccaa gcgtgttccc actggctcct 840
agctctaagt ctacctccgg aggaacagcc gctctgggct gtctggtgaa ggattacttc 900
ccagagcccg tgacagtgtc ttggaactcc ggcgccctga cctctggagt gcatacattt 960
cctgctgtgc tgcagtccag cggcctgtat tccctgtctt ccgtggtgac cgtgccaagc 1020
tcttccctgg gcacccagac atacatctgc aacgtgaatc acaagccctc caatacaaag 1080
gtggacaaga aggtggagcc taagagctgt gataagaccc atacatgccc cccttgtcct 1140
gctccagagc tgctgggcgg accaagcgtg ttcctgtttc cacccaagcc caaggacacc 1200
ctgatgatct ccagaacccc cgaggtgaca tgcgtggtgg tggacgtgtc ccacgaggat 1260
cctgaggtga agtttaactg gtacgtggat ggcgtggagg tgcataatgc caagaccaag 1320
cccagggagg agcagtacaa ttctacctat cgggtggtgt ccgtgctgac agtgctgcac 1380
caggactggc tgaacggcaa ggagtacaag tgcaaggtga gcaataaggc cctgcccgct 1440
cctatcgaga agaccatctc taaggctaag ggccagccaa gggagcccca ggtgtacacc 1500
ctgcctccaa gccgggacga gctgaccaag aaccaggtgt ctctgacatg tctggtgaag 1560
ggcttctacc catccgacat cgccgtggag tgggagagca atggccagcc cgagaacaat 1620
tataagacca caccccctgt gctggacagc gatggctctt tctttctgta cagcaagctg 1680
accgtggata agtctcgctg gcagcagggc aacgtgttta gctgttctgt gatgcatgag 1740
gccctgcaca atcattacac acagaagtcc ctgagcctgt ctcctggcaa g 1791
- 一种重组双功能融合蛋白及应用
- 一种新的重组双功能融合蛋白及其制备和应用
